Sequence Analysis
The sequence analysis is able to find sequences that are common to several or all data sets / scenarios / participants. The sequences can be sequences of AOIs or any similar data, i.e. data that consists of a fixed number of different values.
The sequence analysis does not consider the length of different values, e.g. if the data consists of values AAABBBBCC, the analyzed sequence is ABC.
When analyzing sequences, there are normally two variables one is interested in: Finding long sequences, and finding sequences that appear often. These two objectives are contrary to each other. The sequence analysis node concentrates on finding long sequences. Regarding the number of appearences of a sequence, there is a setting for requiring at least k occurences. But even then the primary objective is finding long sequences, not finding sequences that appear as often as possible. So technically, the sequence analysis finds the n longest common subsequences that appear at least k times in at least x% of the input sequences.
Output
The sequence analysis has two outputs. The first output provides the found sequences themselves. These are also displayed in the preview. The second output outputs a marking on the base data, marking all the occurences of the found sequences, similar to the operation of the sequence search. As with any node that outputs markings, the display of these markings can be (de)activated in the workflow explorer.
Aggregation
The sequence analysis is not able to aggregate, as aggregation does not have a meaningful definition in its context.
Settings
- Column:
- The data column in which the sequence analysis is performed. Only enumerable columns are available for selection.
- Common Subsequence Count:
- The number of common subsequences that should be found. Note, that the result can be a larger number of sequences than given here, as the sequence analysis will always output all results of equal length. E.g. if you set 2 as the common subsequence count, and there is one common subsequence of length 6, but 3 of length 5, the analysis will output all 4 sequences.
- Min Occurence Count Per Sequence:
- How often a subsequence must at least occur per input sequence in order to be a possible result. Assuming that you have 1 data set with 1 scenario and 10 participants, setting this value to 3 means that all found subsequences occur at least 3 times in each participant's data.
- In Min % of Sequences:
Determines in how many sequences a subsequences needs to be found in order to be a possible result. The standard value of 100% means that a subsequence needs to be found in all data set/scenario/participant combinations. Assuming that you have 1 data set with 1 scenario and 10 participants, setting this value to 80% means that a subsequence needs to be found in the data of at least 8 participants in order to be considered for longest common subsequence.
Note, that the number of found results might change when you change this parameter. In the example, with the standard value of 100%, the algorithm might find 8 values of length 5. They will all be displayed, despite the Common Subsequence Count being 3. If now the value is reduced to 80%, the algorithm might find 3 sequences of length 10, that only occur for 8 participants. Then, only those 3 sequences will be displayed, thus effectively reducing the number of found results.
- Input Sequence Kind:
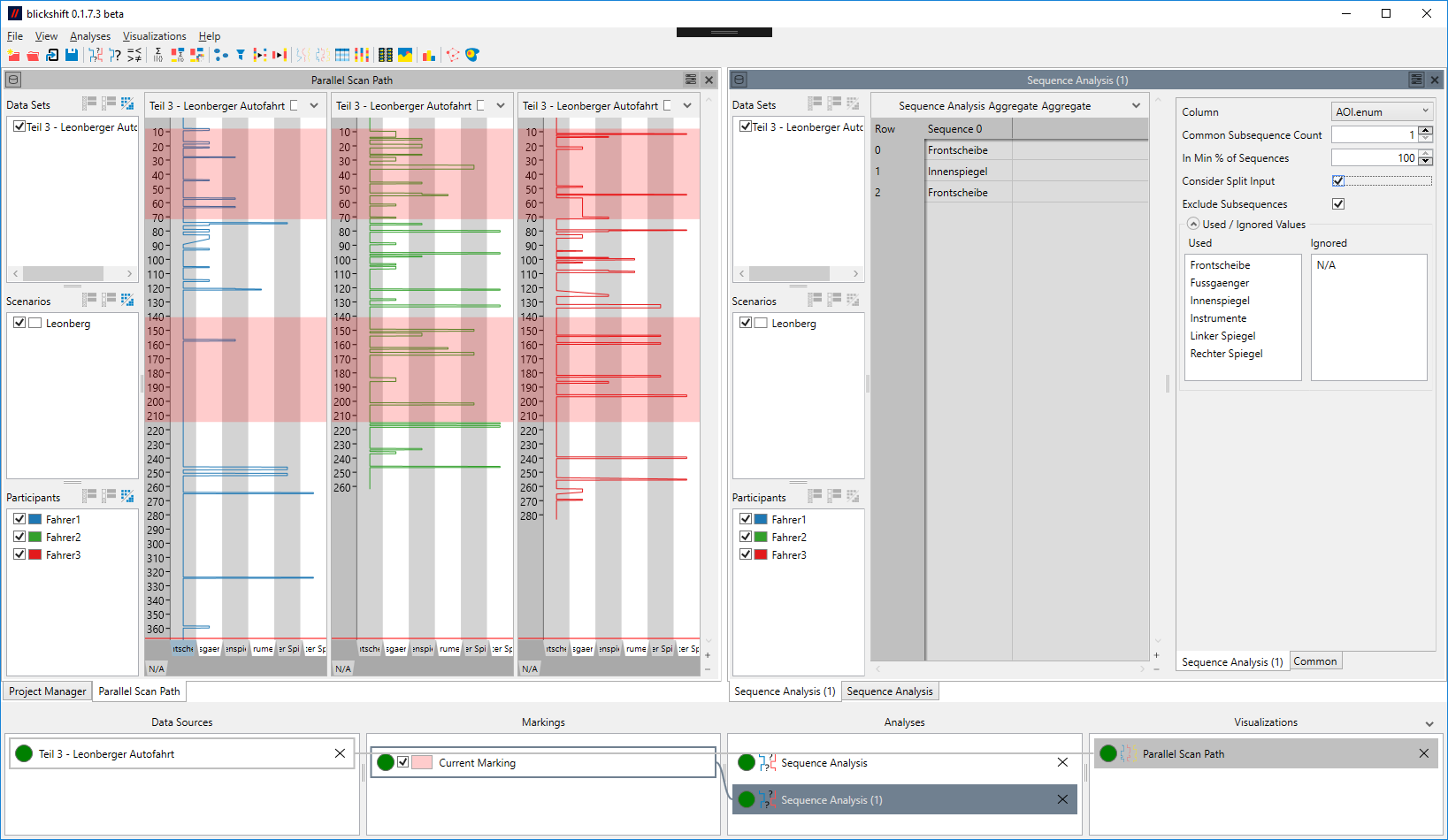
If this option is set to "Section of a Marking", and the input is a split marking, a subsequence must be found not only in each data set/scenario/participant combination, it must also be in each section of the data. If it is set to "Data Table", a subsequence is only required to exist in each data set/scenario/participant combination.
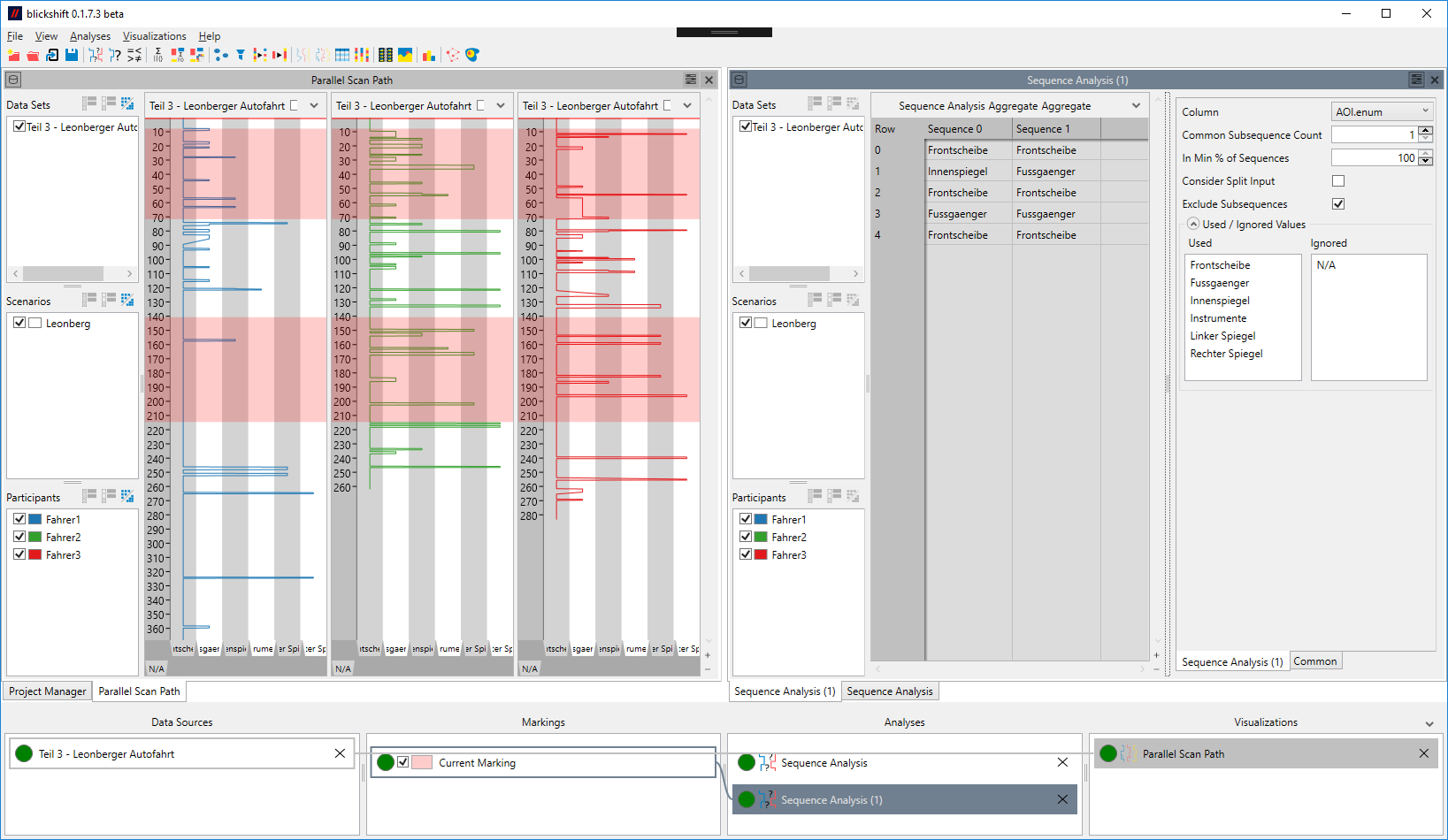
Consider the following example: Figure 1 shows a sequence analysis that analyzes the region marked in the parallel scan path on the left. As it does not consider the split input, two lengthy sequences are found that occur in the data of all participants. Figure 2 shows what happens when it is required that a sequence occurs in every section of the input: Only one short sequence can be found in all marked section. The section marked in the lower half of the blue participant gives an immediate indication of why no longer sequence could be found.
- Split Results:
- Determines whether the marking result on the second output will be a single data set or a different data set for each found sequence. Activating this option can be useful when each found sequence should be analyzed or visualized separately in the following node. Additionally, activating this option makes it possible to determine overlapping found sequences, if the result is shown in a visualization of the base data.